Quick start¶
We assume here that the program is installed.
You can see all available options with:
integron_finder -h
For impatient¶
Go to the directory containing your input file(s), or specify the path to that file and call:
integron_finder mysequences.fst
or:
integron_finder path/to/mysequences.fst
It will perform a search, and outputs the results in a directory called
Results_Integron_Finder_mysequences.
Input and Outputs¶
Inputs¶
integron_finder can take as an input:
- a fasta file
- a multi-fasta file
- many (multi-)fasta files
Outputs¶
By default, integron_finder will output 3 files under Results_Integron_Finder_mysequences:
mysequences.integrons: A file with all integrons and their elements detected in all sequences in the input file.mysequences.summary: A summary file with the number and type of integrons per sequence.integron_finder.out: A copy standard output. The stdout can be silenced with the argument--mute
The amount of log in the standard output can be controlled with --verbose for more or --quiet for less,
and both are cumulative arguments, eg. -vv or -qq.
Other files can be created on demand:
--gbk: Creates a Genbank files with all the annotations found (present in the.integronsfile)--pdf: Creates a simple pdf graphic with complete integrons--split-results: Creates a.integronsa.summaryfile per replicon if the input is a multifasta file.--keep-tmp: Keep temporary files. See Keep intermediate files for more.
For everyone¶
Note
The different options will be shown separately, but they can be used altogether unless otherwise stated.
Thorough local detection¶
This option allows a much more sensitive search of attC sites. It will be slower if integrons are found, but will be as fast if nothing is detected.
integron_finder mysequences.fst --local-max
CALIN detection¶
By default CALIN are reported if they are composed of at least 2 attC sites, in order to avoid false positives. This value was chosen as CALIN with 2 attC sites were unlikely to be false positive. The probability of a false CALIN with at least 2 attC sites within 4kb was estimated between 4.10^-6 and 7.10^-9. However, one can modify this value with the option –calin-threshold and use a lower or higher value depending on the risk one is willing to take:
integron_finder mysequences.fst --calin-threshold 1
Note
If --local-max is called, it will run around CALINs with single attC sites, even if --calin-threshold is 2.
The filtering step is done after the search with local max in that case.
Functional annotation¶
This option allows to annotate cassettes given HMM profiles. As Resfams database is distributed, to annotate antibiotic resistance genes, just use:
integron_finder mysequences.fst --func-annot
IntegronFinder will look in the directory
Integron_Finder-x.x/data/Functional_annotation and use all .hmm files
available to annotate. By default, there is only Resfams.hmm, but one can
add any other HMM file here. Alternatively, if one wants to use a database which
is present elsewhere on the user’s computer without copying it into that
directory, one can specify the following option
integron_finder mysequences.fst --path_func_annot bank_hmm
where bank_hmm is a file containing one absolute path to a hmm file per
line, and you can comment out a line
~/Downloads/Integron_Finder-x.x/data/Functional_annotation/Resfams.hmm
~/Documents/Data/Pfam-A.hmm
# ~/Documents/Data/Pfam-B.hmm
Here, annotation will be made using Pfam-A et Resfams, but not Pfam-B. If a protein is hit by 2 different profiles, the one with the best e-value will be kept.
Search for promoter and attI sites¶
By default integron_finder look for attC sites and site-specific integron integrase,,
If you want to search for known promoters (integrase, Pc-int1 and Pc-int3) and AttI sites
in integrons elements you need to add the --promoter-attI option on the command line.
Keep intermediate results¶
Integrons finder needs some intermediate results to run completely.
It includes notably the protein file in fasta (mysequences.prt), but also the outputs from hmmer and infernal.
A folder containing these outputs is generated for each replicon and have name tmp_<replicon_id>
This directory is removed at the end. You can keep this directory to analyse further each integron_finder steps
with the option --keep-tmp. Using this argument allows you to rerun integron_finder
on the same sequences without redetecting proteins and attC sites. It is useful if one wants to change
clustering parameters, evalues of attC sites, or size of them. Note that it won’t search for new attC sites
so it is better to start with relaxed parameters and then rerun integron_finder with more strict parameters.
See the section for integron diggers for more informations
For each tmp file, there are:
<replicon_id>.fst: a single fasta file with the replicon_name<replicon_id>.prt: a multifasta file with the sequences of the detected proteins.<replicon_id>_intI_table.res: hmm result for the intI hmm profile in tabular format<replicon_id>_intI.res: hmm result for the intI hmm profile<replicon_id>_phage_int_table.res: hmm result for the tyrosine recombinase hmm profile in tabular format<replicon_id>_phage_int.res: hmm result for the tyrosine recombinase hmm profile in tabular format<replicon_id>_attc_table.res: cmsearch result for the attC sites covariance model in tabular format<replicon_id>_attc.res: significant (according toevalue-attc) attC sites aligned in stockholm formatintegron_max.pickle: pickle file sointegron_finderreuse this instead of re-running the local_max part
Topology¶
By default, IntegronFinder assumes that
- your replicon is considered as circular if there is only one replicon in the input file.
- your replicons are considered as linear if there are several replicons in the input file.
However, you can change this default behavior and specify the default topology with options
--circ or --lin:
integron_finder --lin mylinearsequence.fst
integron_finder --circ mycircularsequence.fst
If you have multiple replicon in the input file with different topologies you can specify a topology for each replicon by providing a topology file. The syntax for the topology file is simple:
- one topology by line
- one line start by the seqid followed by ‘circ’ or ‘lin’ for circular or linear topologies.
example:
seq_id_1 circ
seq_id_2 lin
You can also mix the options --circ or --lin with option --topology-file:
integron_finder --circ --topology-file path/to/topofile mysequencess.fst
In the example above the default topology is set to circular. The replicons specified in topofile supersede the default topology.
Warning
However, if the replicon is smaller than 4 x dt
(where dt is the distance threshold, so 4kb by default), the replicon is considered linear
to avoid clustering problem.
The topology used to searching integron is report in the *.integrons file
For big data people¶
Parallelization¶
The time limiting part are HMMER (search integrase) and INFERNAL (search attC sites). So if you have to analyze one or few replicons the user can set the number of CPU used by HMMER and INFERNAL:
integron_finder mysequences.fst --cpu 4
Default is 1.
If you want to deal with a fasta file with a lot of replicons (from 10 to more than thousand) we provide a workflow to parallelize the execution of the data. This mean that we cut the data input into chunks (by default of one replicon) then execute IntegronFinder in parallel on each replicon (the number of parallel tasks can be limited) then aggregate the results in one global summary. The workflow use the nextflow framework and can be run on a single machine or a cluster.
First, you have to install nextflow first, and integron_finder. Then we provide 2 files (you need to download them from the IntegronFinder github repo.)
- parallel_integron_finder.nf which is the workflow itself in nextflow syntax
- nextflow.config which is a configuration file to execute the workflow.
The workflow file should not be modified. Whereas the profile must be adapted to the local architecture.
- The file nextflow.config provide for profiles:
- a standard profile for local use
- a cluster profile
- a standard profile using singularity container
- a cluster profile using singularity container
Warning
On Ubuntu Bionic Beaver (18.04) The default java is not suitable to run nextflow So you have to install another jvm
sudo add-apt-repository ppa:webupd8team/java sudo apt-get update sudo apt-get install oracle-java8-installer
for more details see: https://medium.com/coderscorner/installing-oracle-java-8-in-ubuntu-16-10-845507b13343
so now install nextflow. If you have capsule error like
CAPSULE EXCEPTION: Error resolving dependencies. while processing attribute Allow-Snapshots: false (for stack trace, run with -Dcapsule.log=verbose)
Unable to initialize nextflow environment
install nextflow (>=0.29.0) as follow (change the nextflow version with the last release)
wget -O nextflow http://www.nextflow.io/releases/v0.30.2/nextflow-0.30.2-all
chmod 777 nextflow
for more details see: https://github.com/nextflow-io/nextflow/issues/770#issuecomment-400384617
How to get parallel_integron_finder¶
The release contains the workflow parallel_integron_finder.nf and the nextflow.config at the top level of the archive But If you use pip to install Integron_Finder you have not easily access to them. But they can be downloaded or executed directly by using nextflow.
to download it
nextflow pull gem-pasteur/Integron_Finder
to get the latest version or use -r option to specify a version
nextflow pull -r release_2.0 gem-pasteur/Integron_Finder
to see what you download
nextflow see Integron_Finder
to execute it directly
nextflow run gem-pasteur/Integron_Finder -profile standard --replicons all_coli.fst --circ
or:
nextflow run -r release_2.0 gem-pasteur/Integron_Finder -profile standard --replicons all_coli.fst --circ
standard profile¶
This profile is used if you want to parallelize IntegronFinder on your machine. You can specify the number of tasks in parallel by setting the queueSize value
standard {
executor {
name = 'local'
queueSize = 7
}
process{
executor = 'local'
$integron_finder{
errorStrategy = 'ignore'
cpu=params.cpu
}
}
}
If you installed IntegronFinder with singularity, just uncomment the container line in the script, and set the proper path to the container.
All options available in non parallel version are also available for the parallel one.
except the --outdir which is not available and --replicons option which is specific to the parallelized version.
--replicons allows to specify the path of a file containing the replicons.
A typical command line will be:
./parallel_integron_finder.nf -profile standard --replicons all_coli.fst --circ
Note
Joker as * or ? can be used in path to specify several files as input.
But do not forget to protect the wild card from the shell for instance by enclosing your glob pattern with simple quote.
nextflow run -profile standard parallel_integron_finder.nf --replicons 'replicons_dir/*.fst'
Two asterisks, i.e. **, works like * but crosses directory boundaries.
Curly brackets specify a collection of sub-patterns.
nextflow run -profile standard parallel_integron_finder.nf --replicons 'data/**.fa'
nextflow run -profile standard parallel_integron_finder.nf --replicons 'data/**/*.fa'
nextflow run -profile standard parallel_integron_finder.nf --replicons 'data/file_{1,2}.fa'
The first line will match files ending with the suffix .fa in the data folder and recursively in all its sub-folders. While the second one only match the files which have the same suffix in any sub-folder in the data path. Finally the last example capture two files: data/file_1.fa, data/file_2.fa
More than one path or glob pattern can be specified in one time using comma. Do not insert spaces surrounding the comma
nextflow run -profile standard parallel_integron_finder --replicons 'some/path/*.fa,other/path/*.fst'
The command above will analyze all files ending by .fa in /some/path with .fst extension in other/path
For further details see: https://www.nextflow.io/docs/latest/channel.html#frompath
Note
The option –outdir is not allowed. Because you can specify several replicon files as input, So in this circumstances specify only one name for the output is a none sense.
Note
The options starting with one dash are for nextflow workflow engine, whereas the options starting by two dashes are for integron_finder workflow.
Note
Replicons will be considered linear by default (see above), here we use –circ to consider replicons circular.
Note
If you specify several input files, the split and merge steps will be parallelized.
If you execute this line, 2 kinds of directories will be created.
- One named work containing lot of subdirectories this for all jobs launch by nextflow.
- Directories named Results_Integron_Finder_XXX where XXX is the name of the replicon file. So, one directory per replicon file will be created. These directories contain the final results as in non parallel version.
cluster profile¶
The cluster profile is intended to work on a cluster managed by SLURM. If You cluster is managed by an other drm change executor name by the right value (see nextflow supported cluster )
You can also managed
The number of task in parallel with the executor.queueSize parameter (here 500). If you remove this line, the system will send in parallel as many jobs as there are replicons in your data set.
The queue with process.queue parameter (here common,dedicated)
and some options specific to your cluster management systems with process.clusterOptions parameter
cluster { executor { name = 'slurm' queueSize = 500 } process{ executor = 'slurm' queue= 'common,dedicated' clusterOptions = '--qos=fast' $integron_finder{ cpu=params.cpu } } }
To run the parallel version on cluster, for instance on a cluster managed by slurm, I can launch the main nextflow process in one slot. The parallelization and the submission on the other slots is made by nextflow itself. Below a command line to run parallel_integron_finder and use 2 cpus per integron_finder task, each integron_finder task can be executed on different machines, each integron_finder task claim 2 cpus to speed up the attC sites or integrase search:
sbatch --qos fast -p common nextflow run parallel_integron_finder.nf -profile cluster --replicons all_coli.fst --cpu 2 --local-max --gbk --circ
The results will be the same as describe in local execution.
singualrity profiles¶
If you use the singularity integron_finder image, use the profile standard_singularity. With the command line below nextflow will download parallel_integron_finder from github and download the integron_finder image from the singularity-hub so you haven’t to install anything except nextflow and singularity.
nextflow run gem-pasteur/Integron_Finder -profile standard_singularity --replicons all_coli.fst --circ
You can also use the integron_finder singularity image on a cluster, for this use the profile cluster_singularity.
sbatch --qos fast -p common nextflow run gem-pasteur/Integron_Finder:2.0 -profile cluster_singualrity --replicons all_coli.fst --cpu 2 --local-max --gbk --circ
In the case of your cluster cannot reach the world wide web. you have to download the singularity image
singularity pull --name Integron_Finder shub://gem-pasteur/integron_finder:2.0
the move the image on your cluster modify the nextflow.config to point on the location of the image, and adapt the cluster options (executor, queue, …) to your architecture
cluster_singularity {
executor {
name = 'slurm'
queueSize = 500
}
process {
container = /path/to/integron_finder
queue = 'common,dedicated'
clusterOptions = '--qos=fast'
withName: integron_finder {
cpus = params.cpu
}
}
singularity {
enabled = true
runOptions = '-B /pasteur'
autoMounts = false
}
}
}
then run it
sbatch --qos fast -p common nextflow run ./parallel_integron_finder.nf -profile cluster_singualrity --replicons all_coli.fst --cpu 2 --local-max --gbk --circ
If you want to have more details about the jobs execution you can add some options to generate report:
Execution report¶
To enable the creation of this report add the -with-report command line option when
launching the pipeline execution. For example:
nextflow run ./parallel_integron_finder.nf -profile standard -with-report [file name] --replicons
It creates an HTML execution report: a single document which includes many useful metrics about a workflow execution. For further details see https://www.nextflow.io/docs/latest/tracing.html#execution-report
Trace report¶
In order to create the execution trace file add the -with-trace command line option when launching the pipeline
execution. For example:
nextflow run ./parallel_integron_finder.nf -profile standard -with-trace --replicons
It creates an HTML timeline for all processes executed in your pipeline. For further details see https://www.nextflow.io/docs/latest/tracing.html#timeline-report
Timeline report¶
To enable the creation of the timeline report add the -with-timeline
command line option when launching the pipeline execution. For example:
nextflow run ./parallel_integron_finder.nf -profile standard -with-timeline [file name] --replicons ...
It creates an execution tracing file that contains some useful information about each process executed in your pipeline script, including: submission time, start time, completion time, cpu and memory used. For further details see https://www.nextflow.io/docs/latest/tracing.html#trace-report
For integron diggers¶
Many options are set to prevent false positives. However, one may want higher sensitivity at the expense of having potentially false positives. Ultimately, only experimental experiments will tell whether a given attC sites or integrase is functional.
Also, note that because of how local_max works (ie. around already detected elements), true attC sites
may be found thanks to false attC sites, because false attC sites may trigger local_max around them.
Hence, one may want to use very relaxed parameters first with the --keep-tmp flag to rerun the analysis on
the same data while restrincting the parameters.
Clustering of elements¶
attC sites are clustered together if they are on the same strand and if they
are less than 4 kb apart (-dt 4000 by default). To cluster an array of attC sites and an integron
integrase, they also must be less than 4 kb apart. This value has been
empirically estimated and is consistent with previous observations showing that
biggest gene cassettes are about 2 kb long. This value of 4 kb can be modified
though:
integron_finder mysequences.fst --distance-thresh 10000
or, equivalently:
integron_finder mysequences.fst -dt 10000
This sets the threshold for clustering to 10 kb.
Note
The option --outdir allows you to chose the location of the Results folder (Results_Integron_Finder_mysequences).
If this folder already exists, IntegronFinder will not re-run analyses already done, except functional annotation.
It allows you to re-run rapidly IntegronFinder with a different --distance-thresh value.
Functional annotation needs to re-run each time because depending on the aggregation parameters,
the proteins associated with an integron might change.
Integrase¶
We use two HMM profiles for the detection of the integron integrase. One for tyrosine recombinase and one for a specific part of the integron integrase. To be specific we use the intersection of both hits, but one might want to use the union of both hits (and sees whether it exists cluster of attC sites nearby non integron-integrase…). To do so, use:
integron_finder mysequences.fst --union-integrases
attC evalue¶
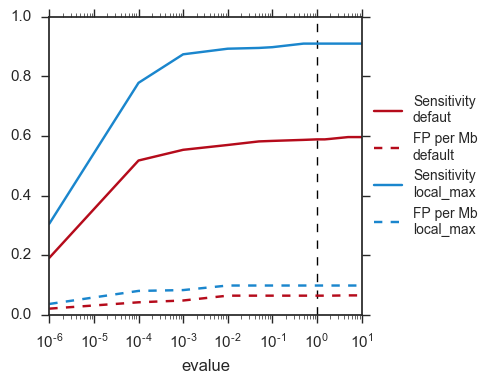
The default evalue is 1. Sometimes, degenerated attC sites can have a evalue above 1 and one may want to increase this value to have a better sensitivity.
integron_finder mysequences.fst --evalue-attc 5
Here is a plot of how the sensitivity and false positive rate evolve as a function of the evalue:
Note
If one wants to have maximum sensitivity, use a high evalue (max is 10), and then integron_finder can be run again on the same data with a lower evalue. It won’t work the other way around (starting with low evalue), as attC sites are not searched again.
attC size¶
By default, attC sites’ size ranges from 40 to 200bp. This can be changed with the --min-attc-size or --max-attc-size parameters:
integron_finder mysequences.fst --min-attc-size 50 --max-attc-size 100
Palindromes¶
attC sites are more or less palindromic sequences, and sometimes, a single attC site can be detected on the 2 strands. By default, the one with the highest evalue is discarded, but you can choose to keep them with the following option:
integron_finder mysequences.fst --keep-palindromes
attC alignements¶
One can get the alignements of attC sites in the temporary files (use --keep-tmp)
to have them. Under Results_Integron_Finder_mysequences/tmp_repliconA/repliconA_attc.res
one can find alignements of attC sites from repliconA, in Stokholm format, where R and L core regions
are aligned with each others:
# STOCKHOLM 1.0
#=GF AU Infernal 1.1.2
ACBA.0917.00019.0001/315102-315161 GUCUAACAAUUC---GUUCAAGCcgacgccgcu.................................................ucgcggcgcgGCUUAACUCAAGC----GUUAGAU
#=GR ACBA.0917.00019.0001/315102-315161 PP ************...******************.................................................***********************....*******
ACBA.0917.00019.0001/313260-313368 ACCUAACAAUUC---GUUCAAGCcgagaucgcuucgcggccgcggaguuguucggaaaaauugucacaacgccgcggccgcaaagcgcuccgGCUUAACUCAGGC----GUUGGGC
#=GR ACBA.0917.00019.0001/313260-313368 PP ************...******************************************************************************************....*******
ACBA.0917.00019.0001/313837-313906 GCCCAACAUGGC---GCUCAAGCcgaccggccagcccu.......................................gcgggcuguccgucgGCUUAGCUAGGGC----GUUAGAG
#=GR ACBA.0917.00019.0001/313837-313906 PP ************...***********************.......................................****************************....*******
#=GC SS_cons <<<<<<<--------<<<-<<<<.....................................................................>>>>>>>---------->>>>>>>
#=GC RF [Rsec=]========[=Lsec=].....................................................................[Lprim]==========[Rprim]
//
Which you can manipulate easily with esl-alimanip tools provided by infernal (the following examples should work if your cmsearch is in your PATH).
You can convert the same alignement in dna alphabet (cmsearch use RNA alphabet):
$ esl-alimanip --dna Results_Integron_Finder_mysequences/tmp_ACBA.0917.00019.0001/ACBA.0917.00019.0001_attc.res
# STOCKHOLM 1.0
#=GF AU Infernal 1.1.2
ACBA.0917.00019.0001/315102-315161 GTCTAACAATTC---GTTCAAGCCGACGCCGCT-------------------------------------------------TCGCGGCGCGGCTTAACTCAAGC----GTTAGAT
#=GR ACBA.0917.00019.0001/315102-315161 PP ************...******************.................................................***********************....*******
ACBA.0917.00019.0001/313260-313368 ACCTAACAATTC---GTTCAAGCCGAGATCGCTTCGCGGCCGCGGAGTTGTTCGGAAAAATTGTCACAACGCCGCGGCCGCAAAGCGCTCCGGCTTAACTCAGGC----GTTGGGC
#=GR ACBA.0917.00019.0001/313260-313368 PP ************...******************************************************************************************....*******
ACBA.0917.00019.0001/313837-313906 GCCCAACATGGC---GCTCAAGCCGACCGGCCAGCCCT---------------------------------------GCGGGCTGTCCGTCGGCTTAGCTAGGGC----GTTAGAG
#=GR ACBA.0917.00019.0001/313837-313906 PP ************...***********************.......................................****************************....*******
#=GC SS_cons <<<<<<<--------<<<-<<<<.....................................................................>>>>>>>---------->>>>>>>
#=GC RF [Rsec=]========[=Lsec=].....................................................................[Lprim]==========[Rprim]
//
You can also convert it to fasta format:
$ esl-alimanip --dna --outformat afa Results_Integron_Finder_mysequences/tmp_ACBA.0917.00019.0001/ACBA.0917.00019.0001_attc.res
>ACBA.0917.00019.0001/315102-315161
GTCTAACAATTC---GTTCAAGCCGACGCCGCT---------------------------
----------------------TCGCGGCGCGGCTTAACTCAAGC----GTTAGAT
>ACBA.0917.00019.0001/313260-313368
ACCTAACAATTC---GTTCAAGCCGAGATCGCTTCGCGGCCGCGGAGTTGTTCGGAAAAA
TTGTCACAACGCCGCGGCCGCAAAGCGCTCCGGCTTAACTCAGGC----GTTGGGC
>ACBA.0917.00019.0001/313837-313906
GCCCAACATGGC---GCTCAAGCCGACCGGCCAGCCCT----------------------
-----------------GCGGGCTGTCCGTCGGCTTAGCTAGGGC----GTTAGAG
The possible outformat are:
- stockholm
- pfam
- a2m
- psiblast
- afa